A Discussion on Issues around Making Data Available Alongside Research
Nicholas Tierney, Monash University
&
Karthik Ram, Berkeley Institute of Data Science, UC Berkeley
Sage Bionetworks
Wednesday 12th February, 2020
"Data! data! data!" he cried impatiently. "I can't make bricks without clay."
--Sherlock Holmes (The Adventure of the Copper Beeches by Sir Arthur Conan Doyle)
This talk represents work that is currently in progress with Karthik Ram:
"A Realistic Guide to Making Data Available Alongside Code to Improve Reproducibility"
Feedback, discussion, questions encouraged throughout talk!
We need data
We need data
We need it to do our job
We need data
We need it to do our job
(though not strictly true for theory, but you get what I mean)
Benefits of Sharing data
🔎 Makes work transparent
✅ Increases trust
🔈 Increases visibility
Independent validation
♻️ Reproducibility
Research isn't often shared
It's often really available to the authors
Rowhani-Farid & Barnett, 2016
Out of 160 randomly sampled BMJ papers:
- 3 included data in the paper
- 7/157 research articles shared their data sets
- For 21 clinical trials bound by the BMJ data sharing policy, 24% shared data
Stodden, Seiler and Ma, 2018
Science (the journal) made clause for Authors to provide data with papers
Authors evaluated reproducibility of 204 papers after clause issued.
Your Turn:
What are the resistance points to data sharing at Sage Bionetworks?
Is data shared publicly?
What is the data sharing process?
Responses to Stodden, Seiler and Ma, 2018
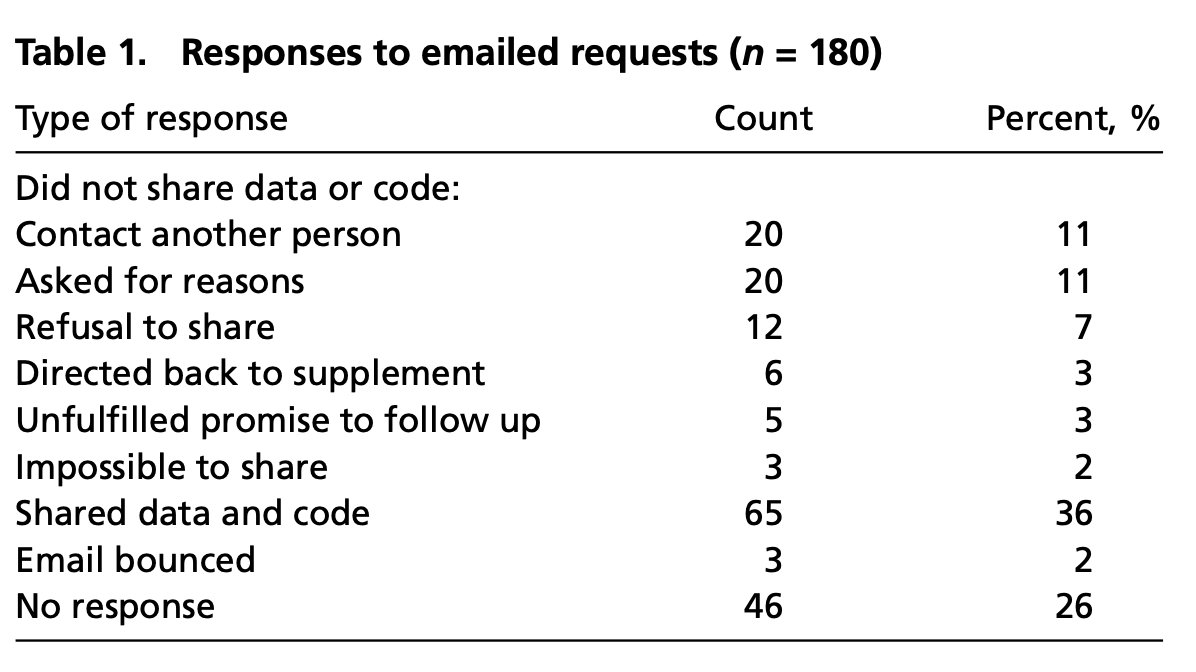
Responses to Stodden, Seiler and Ma, 2018
😿
When you approach a PI for the source codes and raw data, you better explain who you are, whom you work for, why you need the data and what you are going to do with it.
Responses to Stodden, Seiler and Ma, 2018
😢
I have to say that this is a very unusual request without any explanation! Please ask your supervisor to send me an email with a detailed, and I mean detailed, explanation.
Responses to Stodden, Seiler and Ma, 2018
🙍
We do not typically share our internal data or code with people outside our collaboration.
Responses to Stodden, Seiler and Ma, 2018
😿
The code we wrote is the accumulated product of years of effort by [redacted] and myself. Also, the data we processed was collected painstakingly over a long period by collaborators, and so we will need to ask permission from them too.
Responses to Stodden, Seiler and Ma, 2018
😢
Normally we do not provide this kind of information to people we do not know. It might be that you want to check the data analysis, and that might be of some use to us, but only if you publish your findings while properly referring to us.
Responses to Stodden, Seiler and Ma, 2018
😭
Thank you for your interest in our paper. For the [redacted] calculations I used my own code, and there is no public version of this code, which could be downloaded. Since this code is not very user-friendly and is under constant development I prefer not to share this code
Responses to Stodden, Seiler and Ma, 2018
🎉
Our program [redacted] is available here [URL redacted] (documentation and tutorials were included)
Responses to Stodden, Seiler and Ma, 2018
🎉
If you go to [URL redacted], under the publications, I have a link to the gitHub repository. I don’t know if I have all of the raw simulated data, but I certainly have the processed data used to make the plots. What do you need? All of the simulated data could of course be regenerated from the code.
Responses to Stodden, Seiler and Ma, 2018
🎉
Please find attached a .zip file called [redacted].zip that has the custom MATLAB [redacted] analysis code. If you run Masterrunfigureone.m this will generate several panels from the paper.
Responses to Stodden, Seiler and Ma, 2018
🎉
In the next email I will enclose the custom image analysis software. This can also be accessed from [URL redacted] where there is a manual and tutorial.
Sharing data?
Plenty of research says it is important
Fata sharing should be FAIR (findable, Accessible, Interoperable, and Reusable)
These don't precicely tell you how to share data
Why not share data?
There are indeed good reasons to not share data:
Privacy concerns (e.g., human subjects, locations of critically endangered species)
May put the authors at a competitive disadvantage ( but data can be embargoed for reasonable periods of time)
Why not share data?
"If you can't do something right, don't do it"
This ^^ is wrong - you can provide something, even if it is just simulated data.
Sharing data (in most cases) has a net positive benefit
Another way to think about sharing data
Mountain

Another way to think about sharing data
Mountain

Ramp

It should instead be an "on-ramp"
It can feel like a wall or a mountain we need to climb.
These require special tools and knowledge.
An on ramp to sharing data
- Analysis ready data: Final data used in analysis
- README: A Human readable description of the data
- Data dictionary: Human readable dictionary of data contents
- Raw data: The original/first data provided
- Scripts: To clean raw data ready for analysis
- License: How to use and share the data
- Citation: How you want your data to be cited
- Machine readable meta data: Make your data searchable
Analysis ready data: Final data used in analysis
project └── data └── crime.csvAnalysis ready data: Final data used in analysis
project └── data └── crime.csv- Dataset(s) in the form used in analysis.
- Ideally in "tidy data" form.
- Plain-text format,
.csv,.tsv,.txt - Binary formats discouraged
- e.g.,
.rda,.rds,.sav,.dta
README: A Human readable description of the data
project ├── data| └── crime.csv└── README.md- Top level of the data repository, (optionally for each dataset)
- The who, what, when, where, why, of your data.
- Guides user how to understand this directory.
- Handy when no reliable standards
.mdallows you to take advantage ofmarkdown
Data dictionary: Human readable dictionary of data contents
project ├── data│ ├── crime.csv│ └── crime-dictionary.csv └── README.md- Human readable description, context, and structure of the data
- Helps familiarise user with data
- It should contain:
- variable names
- variable labels
- variable codes, and
- special values for missing data
Data Dictionary: Human readable dictionary of data contents
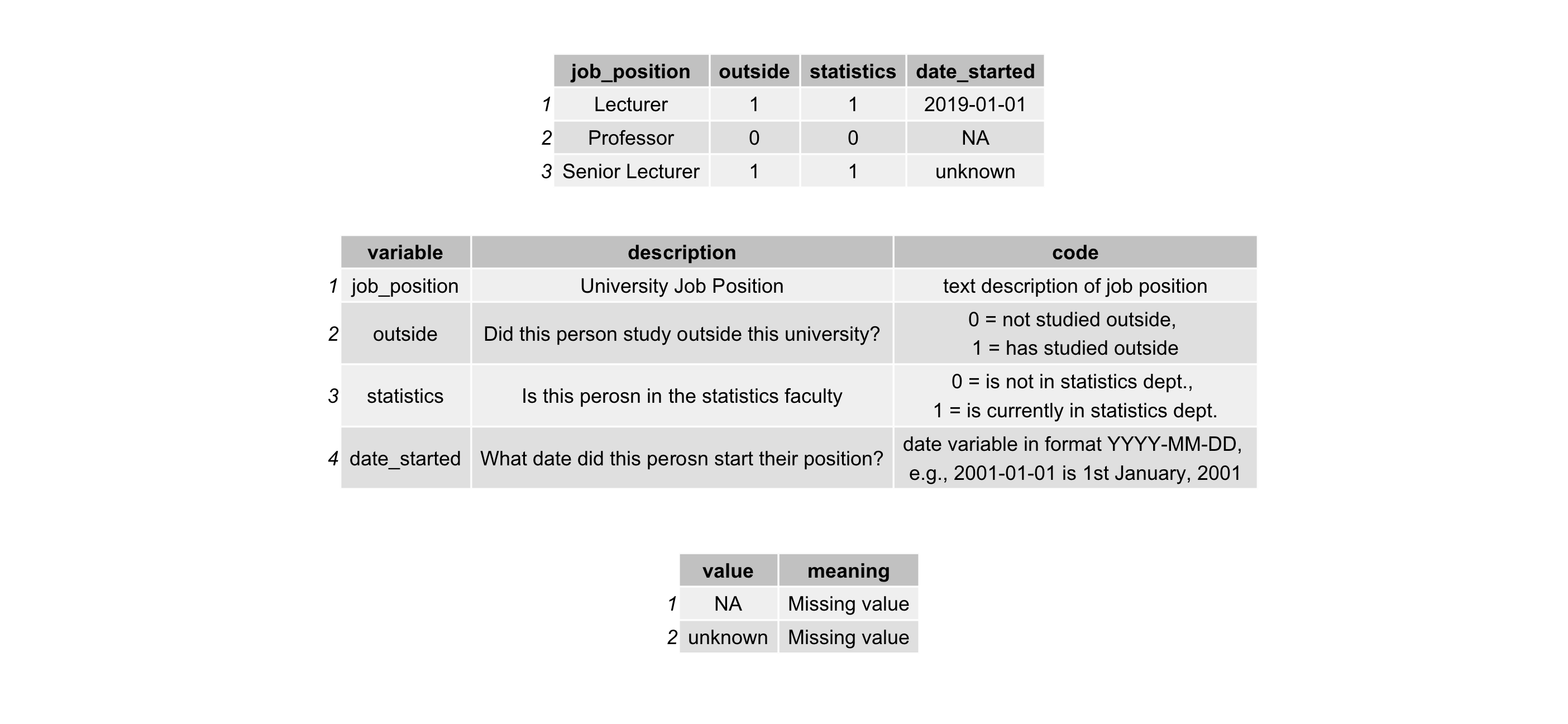
Raw data: The original/first data provided
project ├── data│ ├── crime.csv│ └── crime-dictionary.csv ├── data-raw│ └── crime-raw.dat└── README.md- usually first format of data provided before any tidying or cleaning.
- If the raw data is a practical size to share, it should be shared in a folder called
data-raw. - Should be in the form that was first received, even if it is in binary or some proprietary format.
- Option to include data dictionaries of the raw data can be provided in
data-raw.
Scripts: To clean raw data
project ├── data│ ├── crime.csv│ └── crime-dictionary.csv ├── data-raw│ ├── crime-raw.dat│ ├── clean-crime.R│ └── other-steps.md└── README.md- Code used to clean and tidy the raw data.
clean-crime.R- Ideally involves only scripted languages
- If other practical steps were taken to clean up the data, these should be recorded in a plain text or markdown file.
other-steps.md
License: How to use and share the data
project ├── data│ ├── crime.csv│ └── crime-dictionary.csv ├── data-raw│ ├── crime-raw.dat│ ├── clean-crime.R│ └── other-steps.md├── README.md└── LICENSE- Data + license clearly establishes how everyone to modify, use, and share data.
- Two licenses well suited for data sharing:
- CCBY: enforce attribution and credit required, no warranty.
- CC0: public domain. No ownership or warranty
- Provide LICENSE file with entire license in the top level of directory.
use_cc0_license()use_ccby_license()
Citation: How to cite your data
project ├── data│ ├── crime.csv│ └── crime-dictionary.csv ├── data-raw│ ├── crime-raw.dat│ ├── clean-crime.R│ └── other-steps.md├── README.md (reference DOI)├── CITATION└── LICENSE- A Digital Object Identifier (DOI) uniquely + permanently identifies a digital object ( paper, poster, or software)
- DOIs are minted by repositories like Dryad or Zenodo for free.
- Put the DOI in a reference format like BibTex (zenodo does this for you)
Citation: example
@software{housing-data, author = {Tony Pino, Nicholas Tierney}, title = {njtierney/melb-housing-data: Added LICENSE.md file}, month = feb, year = 2019, publisher = {Zenodo}, version = {1.0.1}, doi = {10.5281/zenodo.2575545}, url = {https://doi.org/10.5281/zenodo.2575545}}Machine readable meta data: Make your data searchable
project ├── data│ ├── crime.csv│ ├── crime-dictionary.csv │ └── metadata│ ├── access.csv│ ├── attributes.csv│ ├── biblio.csv│ ├── creators.csv│ └── dataspice.json├── data-raw│ ├── crime-raw.dat│ ├── clean-crime.R│ └── other-steps.md├── README.md (reference DOI here)├── CITATION└── LICENSE- Helps ensure data types are preserved.
- Allows data to be indexed and searched online with google datasets search using JSON-LD.
- To create appropriate metadata, we recommend metadata generators such as
dataspiceorcodebook - Metadata should be provided in a folder called "metadata", which should be provided for every dataset.
library(dataspice)create_spice(here::here("data"))prep_attributes()prep_access()edit_access()edit_attributes()edit_biblio()edit_creators()write_spice()Actually sharing the data
Now that you've created your data folder, you need to get it somewhere online
Two options to discuss:
- Putting the data online
- Sharing as an R package
Your Turn:
How does Sage Bionetworks distribute data?
Online repositories: Zenodo & Dryad
Zenodo
- Launched in 2013 in a joint collaboration between openAIRE and CERN
- Free, archival location to deposit datasets
- File size limit is 50gb for individual files
- Able to accommodate larger file sizes upon request
Dryad
- The Dryad Digital Repository takes data from any field of research, and perform human quality control and assistance of the data
- Can link data with a journal publication, in exchange for a data publishing fee.
Linking zenodo and GitHub
You can also link Zenodo with Github
Zenodo updates with new DOI at every "release" (Helps avoid managing many moving pieces)
See this article on github, making your code citable (Thanks to Arfon Smith)
Sharing data as an R package
e.g., nycflights13, eechidna, more
Pros
- installable
- documentation
- share data cleaning
- great for R users
Cons
- Size: ! >= 5Mb (CRAN)
- Doesn't help others outside R
Sharing data as an R package
- Most (but not all!) data shared as an R package, or with an R package is for teaching purposes
Sharing data as an R package
- Most (but not all!) data shared as an R package, or with an R package is for teaching purposes
- Sharing data like this allows you to create a "research compendium", with centralised code + paper + computing environment + data.
Sharing data as an R package
- Most (but not all!) data shared as an R package, or with an R package is for teaching purposes
- Sharing data like this allows you to create a "research compendium", with centralised code + paper + computing environment + data.
- See more about this in Karthik Ram's talk: "How To Make Your Data Analysis Notebooks More Reproducible"
Sharing data as an R package
- Most (but not all!) data shared as an R package, or with an R package is for teaching purposes
- Sharing data like this allows you to create a "research compendium", with centralised code + paper + computing environment + data.
- See more about this in Karthik Ram's talk: "How To Make Your Data Analysis Notebooks More Reproducible"
- Note the directory structure based is on R packages
Online "data" journals
- Provides familiar mechanism for citation
- But journals don't yet have a good way to outline how to share data
- Often looks like a "mini paper" with the methods, and isn't always about the data, but about collection methods.
- Can link to a Zenodo or Dryad repository.
Online "data" journals
Some example data journals:
- Nature: Scientific Data
- Data in Brief
- Data
Take homes
- You don't have to do every single thing to publish your data
- Take small steps - get the data somewhere first, add more detail as you go
Future Directions
- Currently working on a proposal for "datadevtools" - a set of developer tools to facilitate sharing data
- These tools can then be used to assess "shareability" of data
Discussion Questions
- Do you curate data?
- What are the common painpoints of curating data / collaborating on data?
- How do you manage data releases?
- What are the resistance points to data sharing?
- Is data shared publicly, is there a process?
- How do you distribute data?
Thanks
- Karthik Ram
- Miles McBain
- Anna Kystalli
- Daniella Lowenberg
- ACEMS International Mobility Programme
- Helmsley Charitable Trust
- Gordon and Betty Moore Foundation
- Sloan Foundation
References
Colophon
- Slides made using xaringan
- Extended with xaringanthemer
- Colours taken + modified from lorikeet theme from ochRe
- Header font is Josefin Sans
- Body text font is Montserrat
- Code font is Fira Mono
- template available: njtierney/njt-talks
Learning more
End.